HIGHLIGHTS
- Changes in the composition of the intestinal microbiota are related to the development of alcoholic liver disease and metabolic-dysfunction associated steatotic liver disease.
- The diversity of the intestinal microbiota was lower in animals with MASLD compared to ALD.
- The structural pattern of the intestinal microbiota was significantly different among the experimental groups.
- Studies are needed to characterize the composition of the intestinal microbiota and metabolome to find new therapeutic strategies.
ABSTRACT – Background –
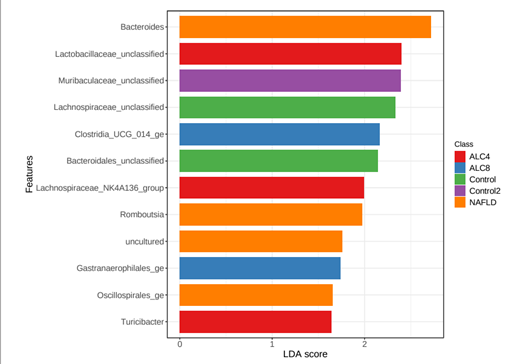
Alcoholic liver disease (ALD) and metabolic-dysfunction associated steatotic liver disease (MASLD) are common, and gut microbiota (GM) is involved with both. Here we compared GM composition in animal models of MASLD and ALD to assess whether there are specific patterns for each disease. Methods – MASLD model— adult male Sprague Dawley rats, randomized into two groups: MASLD-control (n=10) fed a standard diet; MASLD-group (n=10) fed a high-fat-choline-deficient diet for 16 weeks. ALD model— adult male Wistar rats randomized: ALDcontrol (n=8) fed a standard diet and water+0.05% saccharin, ALD groups fed with sunflower seed and 10% ethanol+0.05% saccharin for 4 or 8 weeks (ALC4, n=8; ALC8, n=8). ALC4/8 on the last day received alcoholic binge (5g/kg of ethanol). Afterwards, animals were euthanized, and feces were collected for GM analysis. Results – Both experimental models induced typical histopathological features of the diseases. Alpha diversity was lower in MASLD compared with ALD (p<0.001), and structural pattern was different between them (P<0.001). Bacteroidetes (55.7%), Firmicutes (40.6%), and Proteobacteria (1.4%) were the most prevalent phyla in all samples, although differentially abundant among groups. ALC8 had a greater abundance of the phyla Cyanobacteria (5.3%) and Verrucomicrobiota (3.2%) in relation to the others. Differential abundance analysis identified Lactobacillaceae_unclassified, Lachnospiraceae_NK4A136_group, and Turicibacter associated with ALC4 and the Clostridia_UCG_014_ge and Gastranaerophilales_ge genera to ALC8. Conclusion – In this study, we demonstrated that the structural pattern of the GM differs significantly between MASLD and ALD models. Studies are needed to characterize the microbiota and metabolome in both clinical conditions to find new therapeutic strategies. Keywords – Animal model; alcoholic liver disease; gut microbiota; metabolicdysfunction associated steatotic liver disease; liver disease.
AUTORES
Cássio Marques PERLIN1,2, Larisse LONGO1,2, Rutiane Ullmann THOEN1,2, Carolina URIBE-CRUZ1,2,3 and Mário Reis ÁLVARES-DA-SILVA1,2,4,5